In this post, we learn how to turn a pdf into a structured text document. To this end, we will use a tool called GROBID outputting a corresponding XML document for each pdf. This approach has these advantages over OCR techniques to be
- light-weighted: computation takes a couple of seconds vs. 3-5 minutes with tesseract, a state-of-the-art OCR framework
- easy to use: you just need to parse XML
- storage-efficient: the resulting (TEI) XML file takes just some kB for an entire paper
- REST-ful: GROBID can be run locally or remotely
I’ll conclude with a brief discussion of the TEI format (semi)structuring a PDF and with an application of GROBID.
What is GROBID?
GROBID stands for GeneRation Of Bibliographic Data. It’s a Java tool which transforms a unstructured PDF into a (semi)structured text format. The analysis of scientific publications is the application for GROBID. It can extract scholarly units, such as references, affiliations, authors, DOIs, and abstracts by utilizing machine learning and deep learning.
How to set up GROBID?
- Obtain the latest version via GitHub, Docker or simply pull the latest release by
$ wget https://github.com/kermitt2/grobid/archive/0.5.5.zip && unzip 0.5.5.zip - Build GROBID using gradle
$ ./gradlew clean install - Run the REST service
$ ./gradlew run
which makes the GROBID service available on port 8070.
How to use GROBID
GROBID offers a web user interface to interactively submit pdfs. In addition, you can invoke these services via a REST API. We discuss both how to use the web user interface and use the REST API, via curl and REST clients from GROBID.
To use the web interface, open http://localhost:8070 in your browser. We use this open access publication in the following:
Nathan, R., Getz, W. M., Revilla, E., Holyoak, M., Kadmon, R., Saltz, D., & Smouse, P. E. (2008). A movement ecology paradigm for unifying organismal movement research. Proceedings of the National Academy of Sciences, 105(49), 19052-19059.
I presume this paper is stored in your home directory as papers/nathan_2009_movement_ecology.pdf.
Then, clicking TEI and uploading the pdf should yield the following: 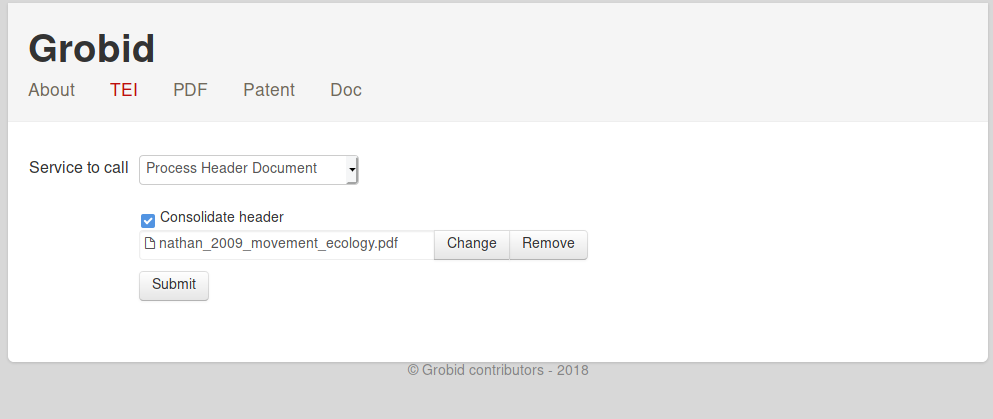 You can decide whether you want to
You can decide whether you want to
- retrieve the paper’s header (
Process Header Document) - get the paper’s entire full text (
Process Fulltext Document) - or obtain just the paper’s references (
Process All References)
After completing generating the xml document, you can either inspect the document or download it:
<?xml version="1.0" encoding="UTF-8"?>
<TEI xmlns="http://www.tei-c.org/ns/1.0" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://www.tei-c.org/ns/1.0 /home/max/code/grobid/grobid-home/schemas/xsd/Grobid.xsd" xmlns:xlink="http://www.w3.org/1999/xlink">
<teiHeader xml:lang="en">
<encodingDesc>
<appInfo>
<application version="0.5.4" ident="GROBID" when="2019-06-28T15:47+0000">
<ref target="https://github.com/kermitt2/grobid">GROBID - A machine learning software for extracting information from scholarly documents</ref>
</application>
</appInfo>
</encodingDesc>
<fileDesc>
<titleStmt>
<title level="a" type="main">A movement ecology paradigm for unifying organismal movement research</title>
</titleStmt>
<publicationStmt>
<publisher>Proceedings of the National Academy of Sciences</publisher>
<availability status="unknown">
<p>Copyright Proceedings of the National Academy of Sciences</p>
</availability>
<date type="published" when="2008-12-09">2008-12-09</date>
</publicationStmt>
<sourceDesc>
<biblStruct>
<analytic>
<author>
<persName xmlns="http://www.tei-c.org/ns/1.0">
<forename type="first">R</forename>
<surname>Nathan</surname>
</persName>
</author>
<author>
<persName xmlns="http://www.tei-c.org/ns/1.0">
<forename type="first">W</forename>
<forename type="middle">M</forename>
<surname>Getz</surname>
</persName>
</author>
<author>
<persName xmlns="http://www.tei-c.org/ns/1.0">
<forename type="first">E</forename>
<surname>Revilla</surname>
</persName>
</author>
<author>
<persName xmlns="http://www.tei-c.org/ns/1.0">
<forename type="first">M</forename>
<surname>Holyoak</surname>
</persName>
</author>
<author>
<persName xmlns="http://www.tei-c.org/ns/1.0">
<forename type="first">R</forename>
<surname>Kadmon</surname>
</persName>
</author>
<author>
<persName xmlns="http://www.tei-c.org/ns/1.0">
<forename type="first">D</forename>
<surname>Saltz</surname>
</persName>
</author>
<author>
<persName xmlns="http://www.tei-c.org/ns/1.0">
<forename type="first">P</forename>
<forename type="middle">E</forename>
<surname>Smouse</surname>
</persName>
</author>
<title level="a" type="main">A movement ecology paradigm for unifying organismal movement research</title>
</analytic>
<monogr>
<title level="j" type="main">Proceedings of the National Academy of Sciences</title>
<title level="j" type="abbrev">Proceedings of the National Academy of Sciences</title>
<idno type="ISSN">0027-8424</idno>
<idno type="eISSN">1091-6490</idno>
<imprint>
<publisher>Proceedings of the National Academy of Sciences</publisher>
<biblScope unit="volume">105</biblScope>
<biblScope unit="issue">49</biblScope>
<biblScope unit="page" from="19052" to="19059"/>
<date type="published" when="2008-12-09"/>
</imprint>
</monogr>
<idno type="DOI">10.1073/pnas.0800375105</idno>
<note type="submission">Edited by James H. Brown, University of New Mexico, Albequerque, NM, and approved June 25, 2008 (received for review March 13, 2008)</note>
</biblStruct>
</sourceDesc>
</fileDesc>
<profileDesc>
<abstract>
<p>Movement of individual organisms is fundamental to life, quilting our planet in a rich tapestry of phenomena with diverse implications for ecosystems and humans. Movement research is both plentiful and insightful, and recent methodological
advances facilitate obtaining a detailed view of individual movement. Yet, we lack a general unifying paradigm, derived from first principles, which can place movement studies within a common context and advance the development of a mature scientific
discipline. This introductory article to the Movement Ecology Special Feature proposes a paradigm that integrates conceptual, theoretical, methodological, and empirical frameworks for studying movement of all organisms, from microbes to trees to
elephants. We introduce a conceptual framework depicting the interplay among four basic mechanistic components of organismal movement: the internal state (why move?), motion (how to move?), and navigation (when and where to move?) capacities of the
individual and the external factors affecting movement. We demonstrate how the proposed framework aids the study of various taxa and movement types; promotes the formulation of hypotheses about movement; and complements existing biomechanical,
cognitive, random, and optimality paradigms of movement. The proposed framework integrates eclectic research on movement into a structured paradigm and aims at providing a basis for hypothesis generation and a vehicle facilitating the understanding
of the causes, mechanisms, and spatiotemporal patterns of movement and their role in various ecological and evolutionary processes. ''Now we must consider in general the common reason for moving with any movement whatever.''
(Aristotle, De Motu Animalium, 4th century B.C.) motion capacity navigation capacity migration dispersal foraging</p>
</abstract>
</profileDesc>
</teiHeader>
<text xml:lang="en"></text>
</TEI>
This document contains already a lot of information about the publication! So what is TEI? TEI stands for Text Encoding Initiative. As its name already hints, TEI is a standard guiding how to encode digital texts. I’ll cover parsing TEI files in another post.
RESTful GROBID
If you have many articles, it becomes handy to output multiple XML documents at the same time in a batch process. To do so, we need to invoke GROBID’s REST API to automate this process. First, let’s obtain the same XML by using curl:
$ curl -v --form input=@./nathan_2009_movement_ecology.pdf localhost:8070/api/processHeaderDocument
Of course, it’s possible to write your own client to submit such requests. However, GROBID already offers clients in Java, Python and JavaScript to process multiple pdfs stored in a folder at the same time. Let’s use the Python client:
$ python3 grobid-client.py --n 3 --input ~/papers --output ~/tei_papers processFulltextDocument
The command will concurrently generate TEI xml documents for all PDFs in ~/papers. The output of this call is stored in directory ~/tei_papers. We can adjust how many threads we want to use by the parameter --n 3.
TEI XML
In the above example, we got a glimpse on how a header for TEI XML looks like. But how is a TEI XML structured if we extract the entire document? Let’s have a look at a stub without any information filled in:
<?xml version="1.0" encoding="UTF-8"?>
<TEI xmlns="http://www.tei-c.org/ns/1.0" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://www.tei-c.org/ns/1.0 /home/max/code/grobid/grobid-home/schemas/xsd/Grobid.xsd" xmlns:xlink="http://www.w3.org/1999/xlink">
<teiHeader xml:lang="en">
...
</teiHeader>
<text xml:lang="en">
<body>
...
</body>
<back>
...
</back>
</text>
</TEI>
teiheadercontains infos about publication date, authors, journal, doi and abstractbodyencompasses the main text including figures and tablesbackanything else like acknowledgments, funding or supplementary materials.
‘Nuff said about data format, how about applications? Let’s have a look where GROBID has been already applied.
Application of GROBID: RobotReviewer
Systematic reviews have an important role in clinical trials. One aspect of such reviews is the risk of bias assessment in form of the PICO (population, intervention, comparators and outcomes) scheme. Automatic reports compiled from publications can be a help to identify how individual papers fall into a PICO scheme. RobotReviewer analyzes such trail characteristics and generates an evidence report.
To generate text documents from PDFs, RobotReviewer internally uses GROBID.
